-
Notifications
You must be signed in to change notification settings - Fork 10
RunKallisto
homonecloco edited this page Oct 22, 2015
·
2 revisions
expVIP can run Kallisto and load the tpm and counts to the database. The only requirement is to run kallisto index on the transcriptome reference.
- Double click on
run_kallisto.sh
- Click on
Execute on terminal
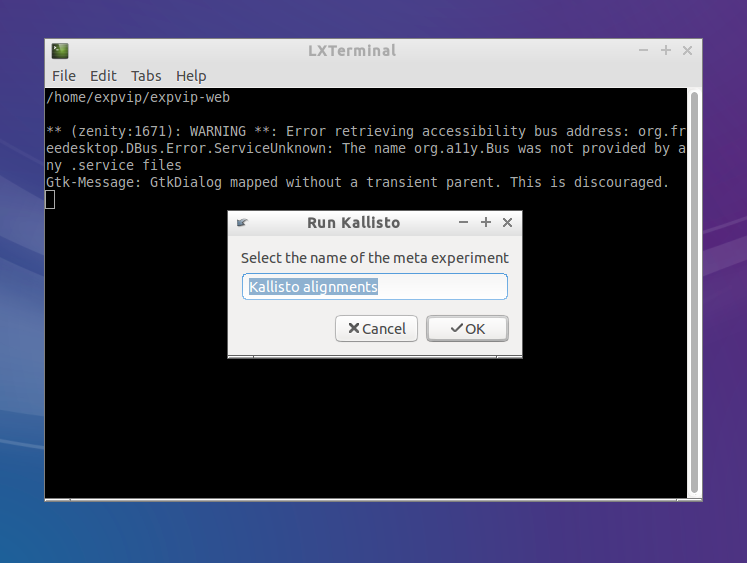
- Give a name to the set of mappings to be grouped. All mappings done with the same reference and preference should have the same name.
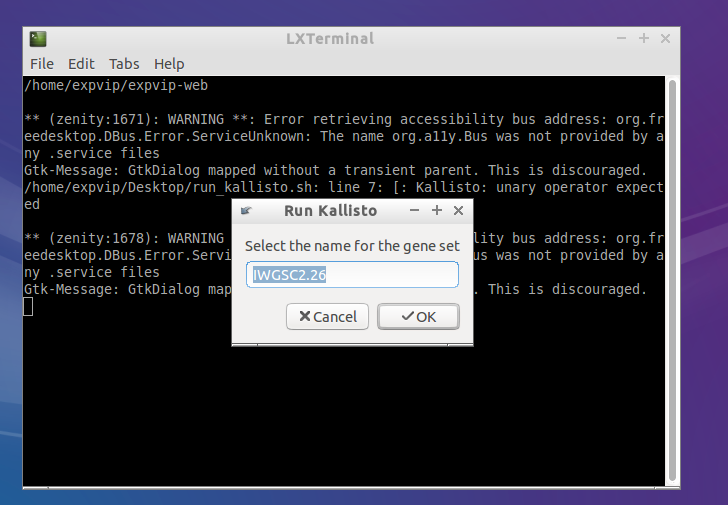
- Get the name of the reference. This name must be the same used when loading the metadata
- Select a folder with the reads. The reads must be paired reads. The folder name must be the same as the
accessionused on the metadata.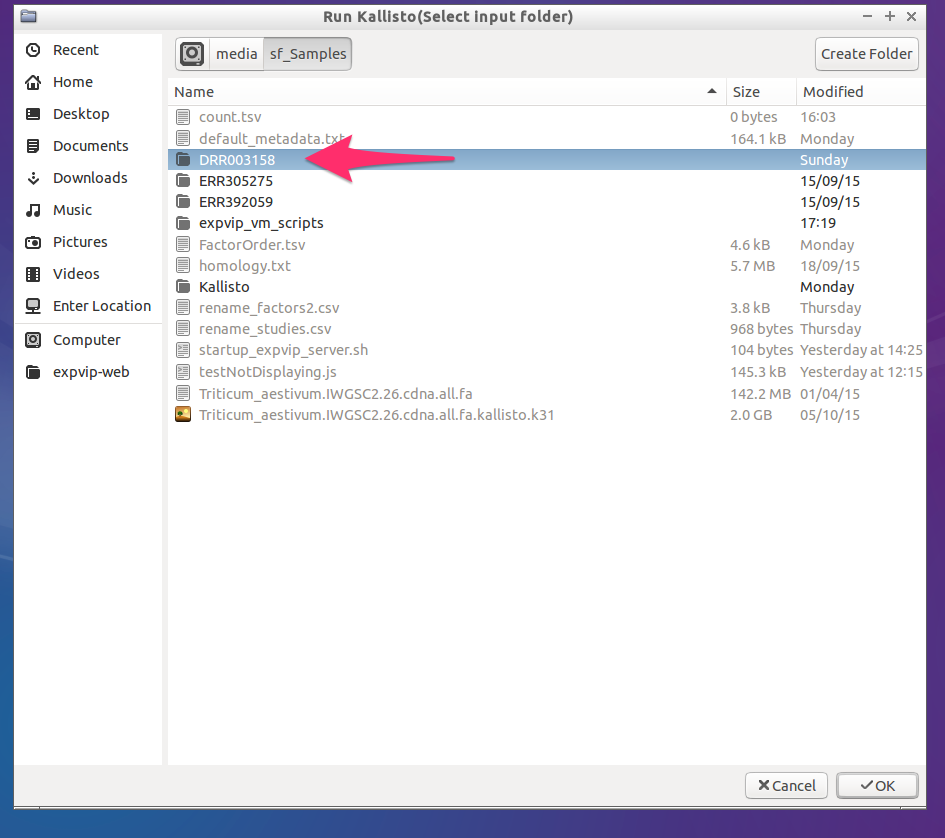
- Select the kallisto index
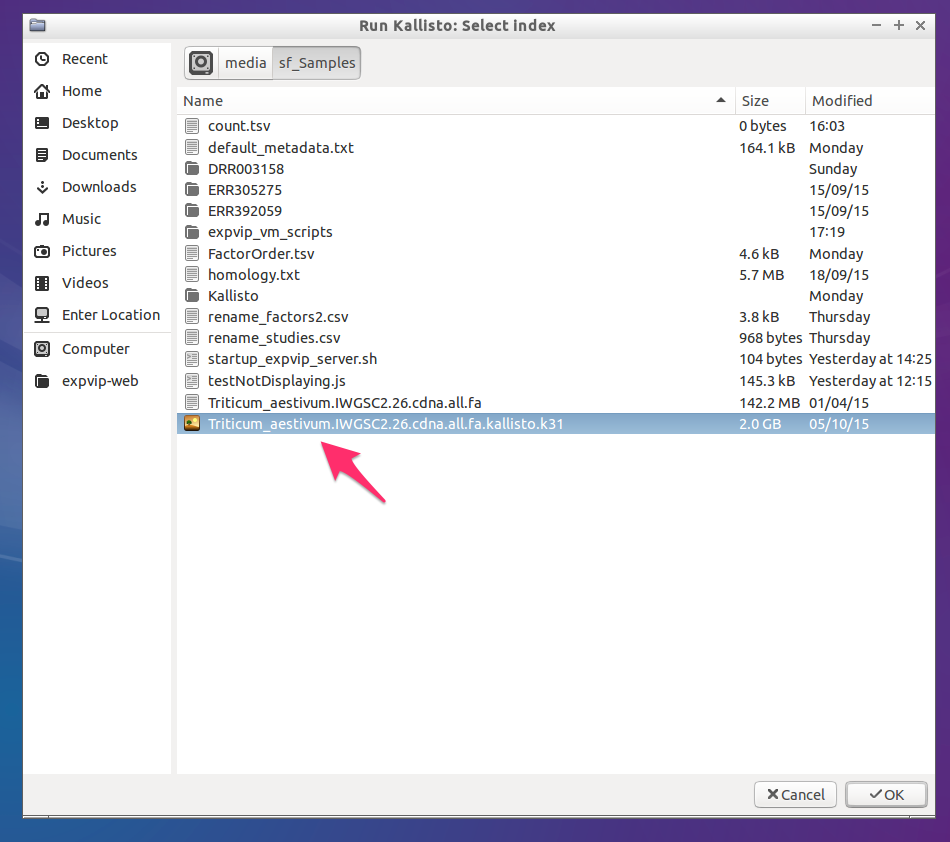
- Wait for Kallisto to run and load the data
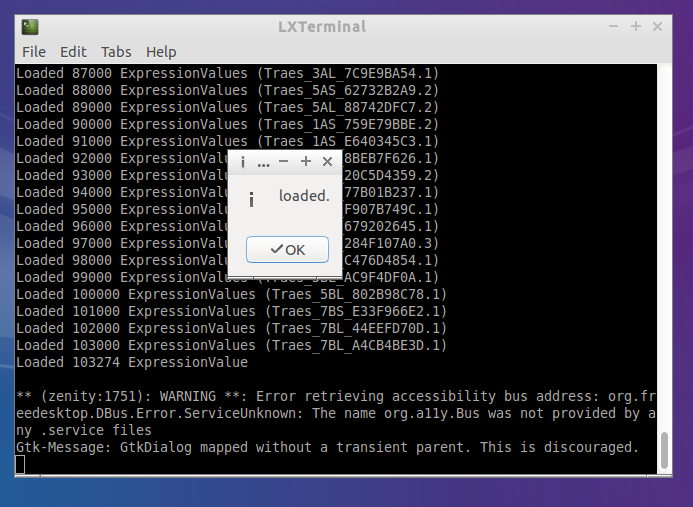
You can reepeat this with all the samples or you can use the batch load.
kallisto:runAndStorePaired[kallistoIndex,input_folder,metaExperimentName,geneSetName]Where metaExperimentName is the name of the group of alignments under the same conditions and ```geneSetName`` is the name of the reference.