- 1. Introduction
- 2. Installation
- 3. Generating the documentation locally
- 4. IBSI Standardization
- 5. Acknowledgement
- 6. Authors
- 7. Statement
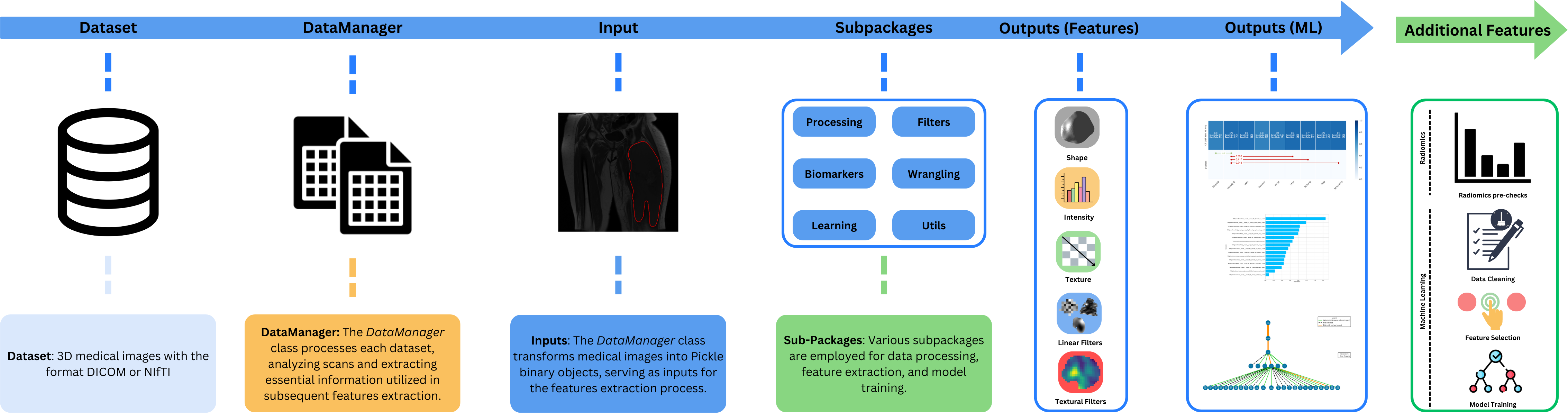
This is an open-source python package for processing and extracting features from medical images. It facilitates the medical-images processing and computation of all types of radiomic features as well as the reproducibility of the different analysis. This package has been standardized with the IBSI norms.
The MEDimage package requires python 3.8 or more to be run. If you don't have it installed on your machine, follow the instructions here.
You can easily install the MEDimagepackage from PyPI using:
pip install MEDimage
For more installation options (conda, poetry...) check out the installation documentation.
We used sphinx to create the documentation for this project and you check it out in this link. But you can generate and host it locally by compiling the documentation source code using:
cd docs
make clean
make html
Then open it locally using:
cd _build/html
python -m http.server
We have created many tutorials for the different functionalities of the package. More details can be found in the documentation
The image biomarker standardization initiative (IBSI) is an independent international collaboration which works towards standardizing the extraction of image biomarkers from acquired imaging. The IBSI therefore seeks to provide image biomarker nomenclature and definitions, benchmark data sets, and benchmark values to verify image processing and image biomarker calculations, as well as reporting guidelines, for high-throughput image analysis. We have participated in this collaboration with our package to make sure it respects the international nomenclatures and definitions The participation was separated to two chapters:
-
The IBSI chapter 1 was initiated in September 2016, and it reached completion in March 2020 and is dedicated to the standardization of commonly used radiomic features. We have created jupyter notebooks and made it available for the users to run the IBSI tests themselves. The tests are an interactive Colab notebooks and is directly accessible here:
-
The IBSI chapter 2 was launched in June 2020 and still in progress. It is dedicated to the standardization of commonly used imaging filters in radiomic studies. We have created jupyter notebooks and made it available for the users to run the IBSI tests themselves and validate image filtering and image biomarker calculations from filter response maps. The tests are an interactive Colab notebooks and is directly accessible here:
Our team UdeS (a.k.a. Université de Sherbrooke) has already submitted the benchmarked values to the IBSI uploading website and it can be found here: IBSI upload page.
Miscellaneous
You can avoid the next steps (jupyter installation and environment set up) if you installed the package using conda or poetry according to the documentation.
You can view & run the tests locally by installing the Jupyter Notebook application on your machine:
python -m pip install jupyter
Then add the installed medimage environment to the jupyter notebook kernels using:
python -m ipykernel install --user --name=medimage
Then access the IBSI tests folder using:
cd notebooks/ibsi/
Finally, launch jupyter notebook to navigate through the IBSI notebooks using:
jupyter notebook
MEDimage is an open source package that welcome any contribution and feedback. We wish that this package could serve the growing radiomics research community by providing a flexible as well as ibsi standardized tool to reimplement existing methods and develop their own new methods.
- MEDomics: MEDomics consortium.
This package is part of https://github.com/medomics, a package providing research utility tools for developing precision medicine applications.
Copyright (C) 2023 MEDomics consortium
GPL3 LICENSE SYNOPSIS
Here's what the license entails:
1. Anyone can copy, modify and distribute this software.
2. You have to include the license and copyright notice with each and every distribution.
3. You can use this software privately.
4. You can use this software for commercial purposes.
5. If you dare build your business solely from this code, you risk open-sourcing the whole code base.
6. If you modify it, you have to indicate changes made to the code.
7. Any modifications of this code base MUST be distributed with the same license, GPLv3.
8. This software is provided without warranty.
9. The software author or license can not be held liable for any damages inflicted by the software.
More information on about the LICENSE can be found here