-
Notifications
You must be signed in to change notification settings - Fork 9
Exercise C: Warping Studies
Once we have subtracted estimated backgrounds from the data, we are left with a selected event distribution in reconstructed variables. The migration matrix we found with the event loop maps our best estimate of true variables to reconstructed variables, so we want to use its inverse for unfolding. Unfortunately, experimental migration matrices are almost never invertible because of effects like smearing during reconstruction and statistical fluctuations in the Monte Carlo. Regularization procedures use extra information from our Monte Carlo simulation to find a minimally biased estimate of the inverse of the migration matrix.
But suppose that our Monte Carlo simulation predicts the wrong event distribution. It might not predict enough 2p2h interactions for example. Then, unfolding our background-subtracted data could further bias our published result away from the physics process we're trying to measure.
Warping studies look for this effect and help us figure out how little regularization bias we can get away with. We change the "fake data", generated as a "warped" Monte Carlo simulation, in ways that we suspect could better simulate some aspect of the natural process we're measuring and unfold it with our nominal migration matrix. We then perform a chi squared test between the unfolded "fake data" and the warped efficiency numerator. As we increase the amount of regularization, measured by the number of times the iterative d'Agostini algorithm is applied, we want this chi squared statistic to approach the number of degrees of freedom (number of bins).
We want to choose the minimum number of d'Agostini iterations that produces an acceptable chi squared statistic across multiple warped models. It's also desirable for the chi squared statistic to be stable with number of iterations around the number we pick. A tool called from PlotUtils TransWarpExtractor produces a chi squared versus iterations histogram like this:
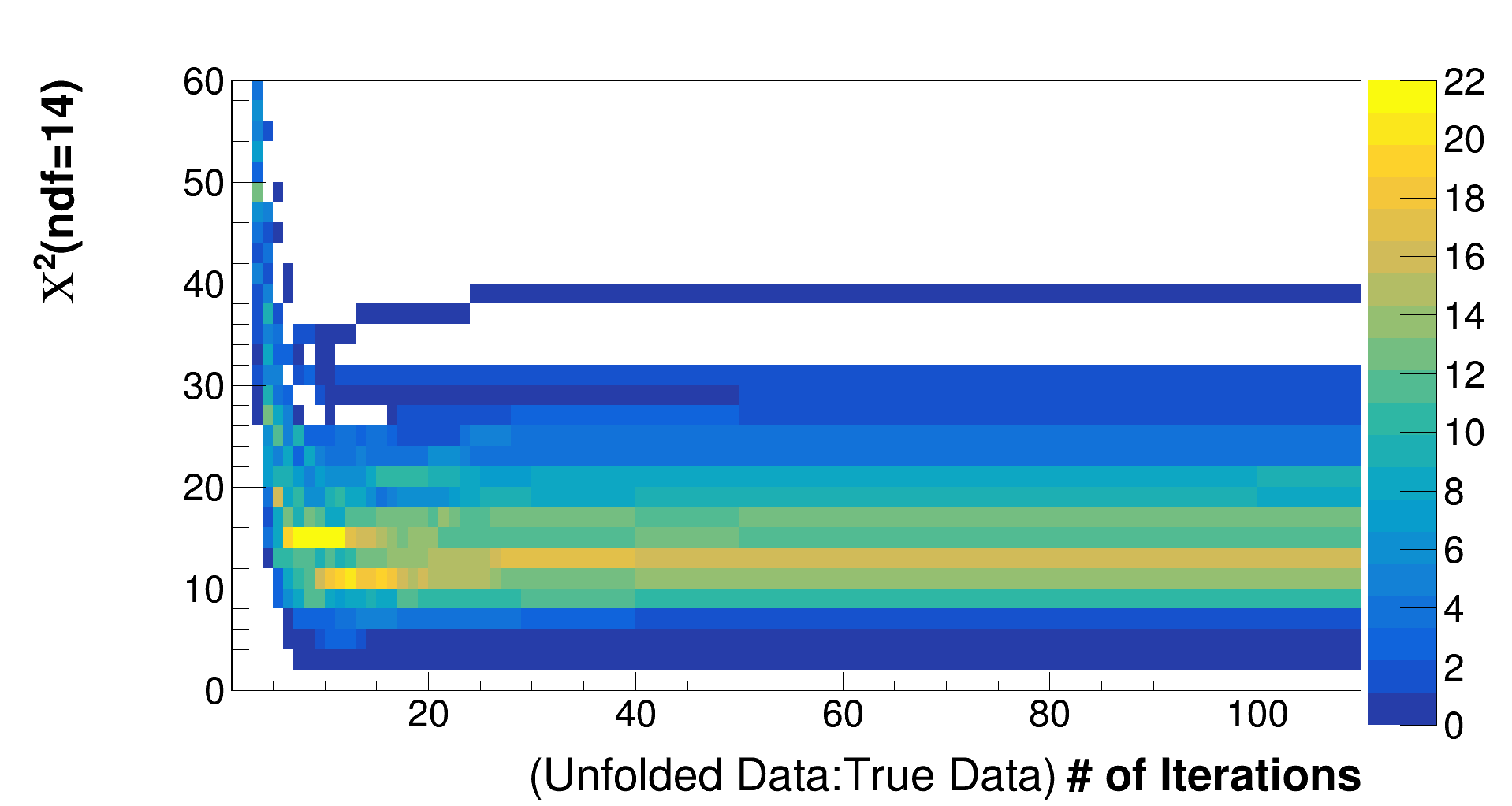
The chi2 is often pretty bad with very few iterations. That's just an indication that the warp is indeed very different from the CV "fake data". So I zoomed in on the y axis.
There are multiple entries per bin because TransWarpExtractor also simulates (Poisson-distributed) statistical fluctuations on each bin. We can also use the truncated mean chi squared (a TProfile in the same TDirectory), like the plot below, to choose a number of iterations for a given warp.

Finally, since the statistical throws make the variance in the mean chi squared depend on total number of events, we need a standard for the sample size used in warping studies. Our standard is 12e20 Monte Carlo POT to match the data exposure. We don't have time to process 12e20 MC POT during MINERvA101 (although FermiGrid might help), so this tutorial is only a first look at a warping study for an inclusive analysis.
- Turn the 2p2h tune off. This is a very aggressive tune that most results violently disagree with. But the inclusive analysis is so generic that it can survive this test.
- Change our pion model. MnvTunev2, the successor to MnvTunev1, has a pion-based reweight that we'll use. The low Q2 pion suppression reweight is an empirical correction for the data/MC discrepancy we see in our coherent pion analysis.
- Change our DIS model. In the Low Energy 2D inclusive result, Amy pointed out that DIS dominates in the high pz, high pT regions where the data and MC disagree most violently. Our colleagues from Aligarh Muslim University had just developed a new model for neutrino Deep Inelastic Scattering that we compared to MnvTunev1. We're going to reweight GENIE to this AMU model to simulate different DIS kinematics in nature.
Modify runEventLoop to produce warped "fake data". Look for the Model object set up in main() and add/remove Reweighters to set up Your Warps. TransWarpExtractor only uses the CV migration matrix, so use runEventLoop with systematics turned off for the warps to make sure you can finish this exercise in time.
The runTransWarp.sh script maps runEventLoop's histograms to TransWarpExtractor's command line arguments. Run it like runTransWarp.sh runEventLoopMC.root warpedMC_AMUDIS.root. It will produce a file named Warping_warpedMC_AMUDIS.root each time it is run. Open these files like this:
root -l Warping_warpedMC_AMUDIS.root
TBrowser tb; //Graphical display of directories, histograms, and other ROOT objects
Now, look at the truncated mean chi2 versus iteration histograms in directory Chi2_Iteration_Dists which are named h_chi2_modelData_trueData_iter_chi2 and choose the minimum number of iterations for each warp. We usually publish a result using the largest minimum number of iterations among 3-4 warps.
Look at one heat map, like the example on this page, for each warp.
- Estimate the iteration where the mean chi squared starts to level off. In the example on this page, I think that happens around 6 iterations.
- We don't want the rough standard deviation in chi squared to be too large at the iteration we choose. In this example, I concluded that the deviation isn't too large after about 6 iterations because the slightly green region spans less than the number of degrees of freedom.
Conclusions for each warp:
| Warp | iterations | comment |
|---|---|---|
| AMU DIS | 6 | good |
| No 2p2h Tune | 8 | good |
| Pion Supp. | 10 | chi squared high |
The chi squared statistics for this analysis only get slightly better at higher number of iterations and worse at very low number of iterations. So we choose to unfold the final result to 10 iterations.
You can see how David implemented the warps to solve this exercise on the branches: warp_2p2hTuneOff, warp_AMUDISTuneOn, and warp_lowQ2PiTuneOn. Remember that you can temporarily make your working area look like one of these branches (and always be able to go back!) with git checkout warp_lowQ2PiTuneOn. You can imagine that changing the source code of your analysis just to do a warping study is kind of error-prone though. Most mature analyses have flags or even lists of reweights they can apply without changing any code.
If you didn't get your event loop running well enough to pass the closure test, you can look at example warping study results using these files instead: AMU DIS, 2p2h tune off, and low Q2 pion suppression