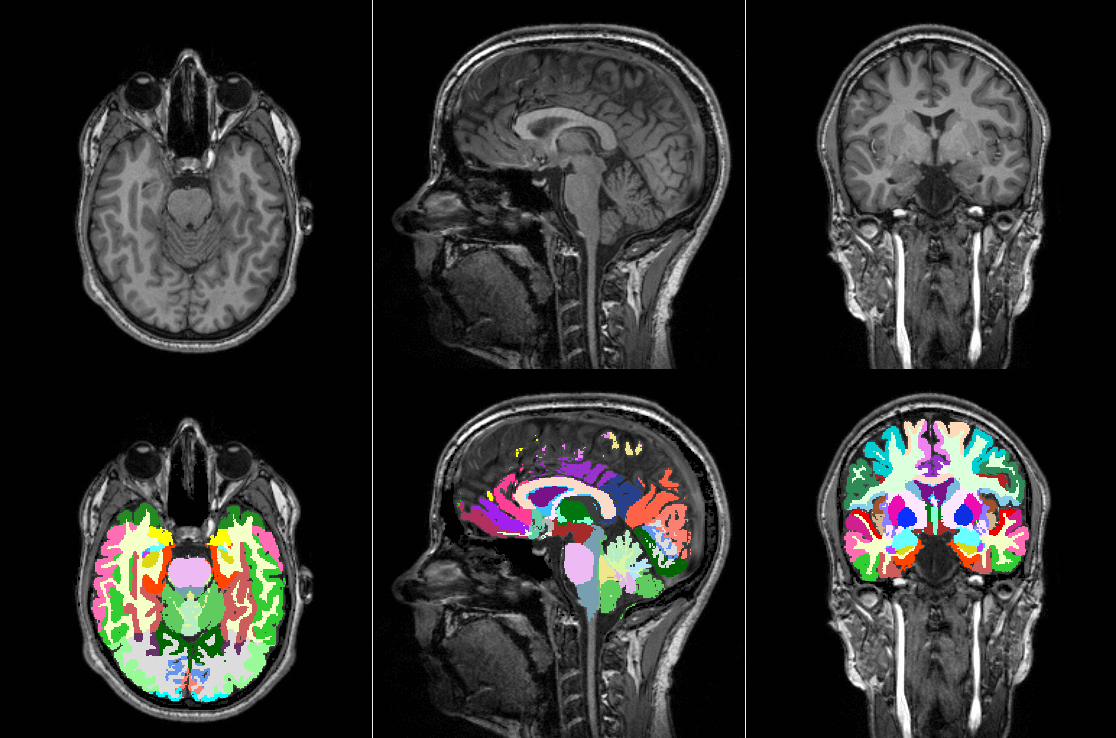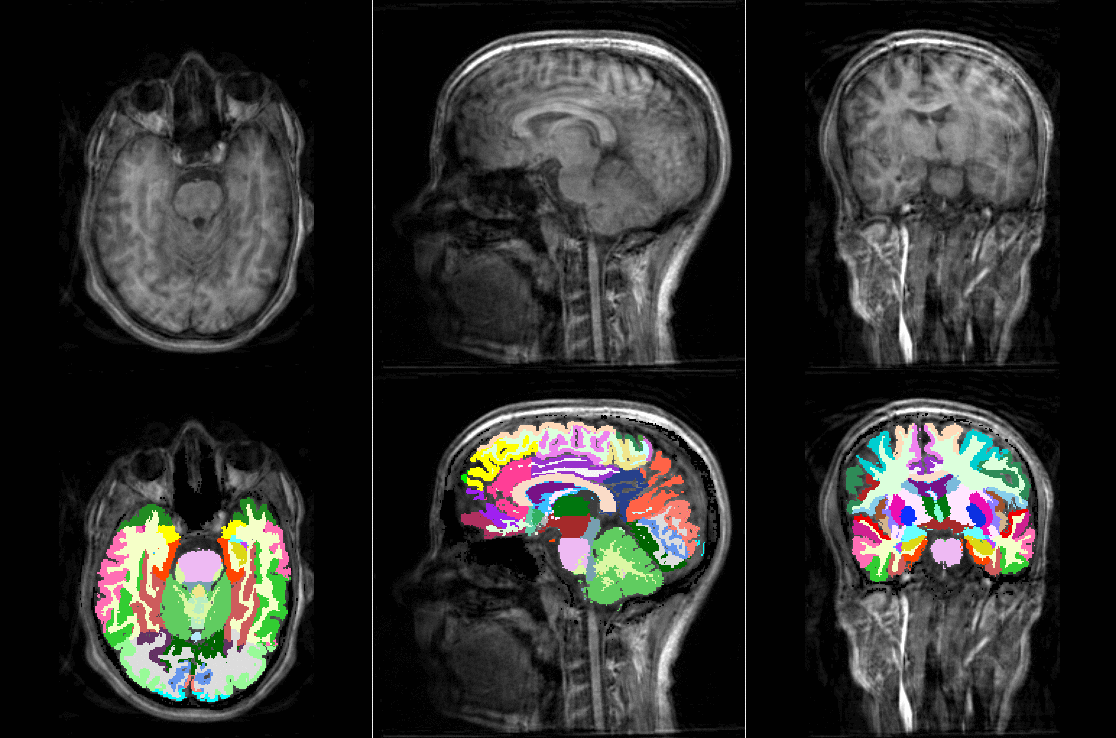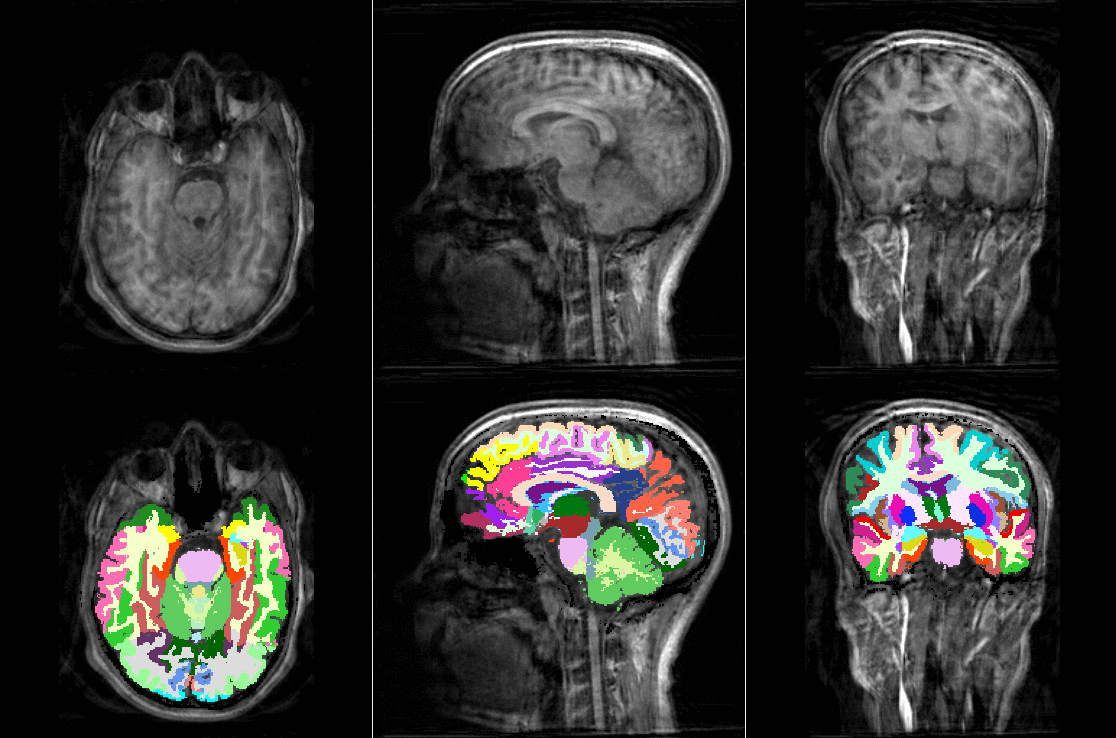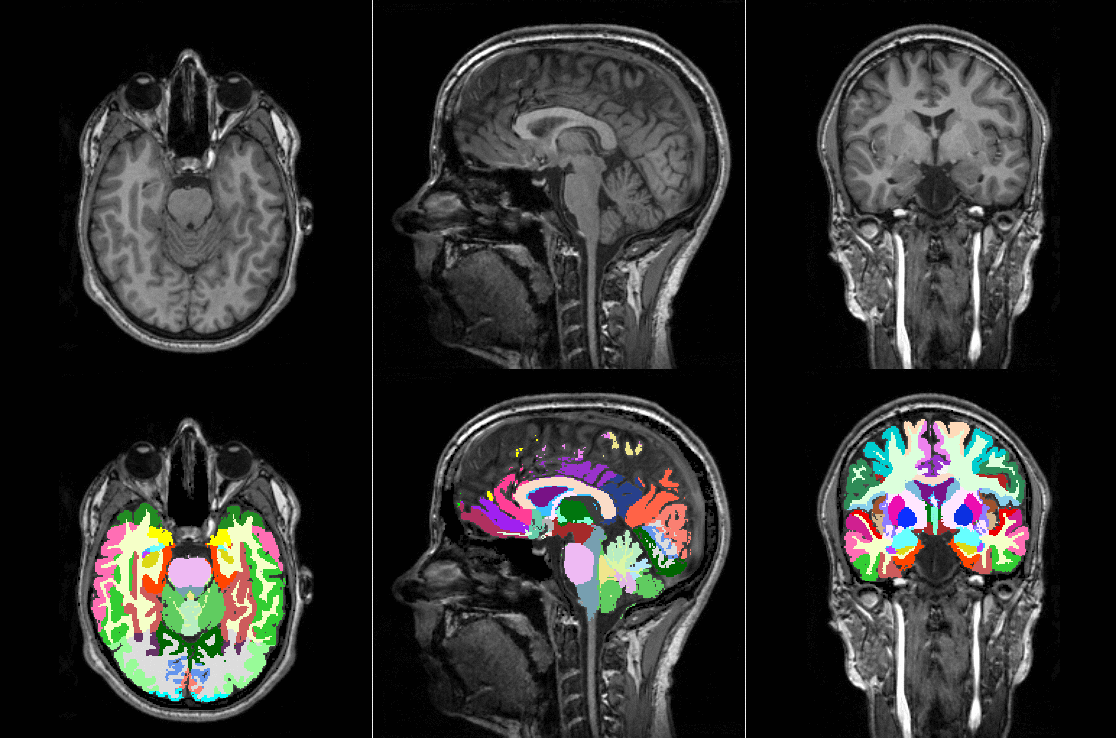
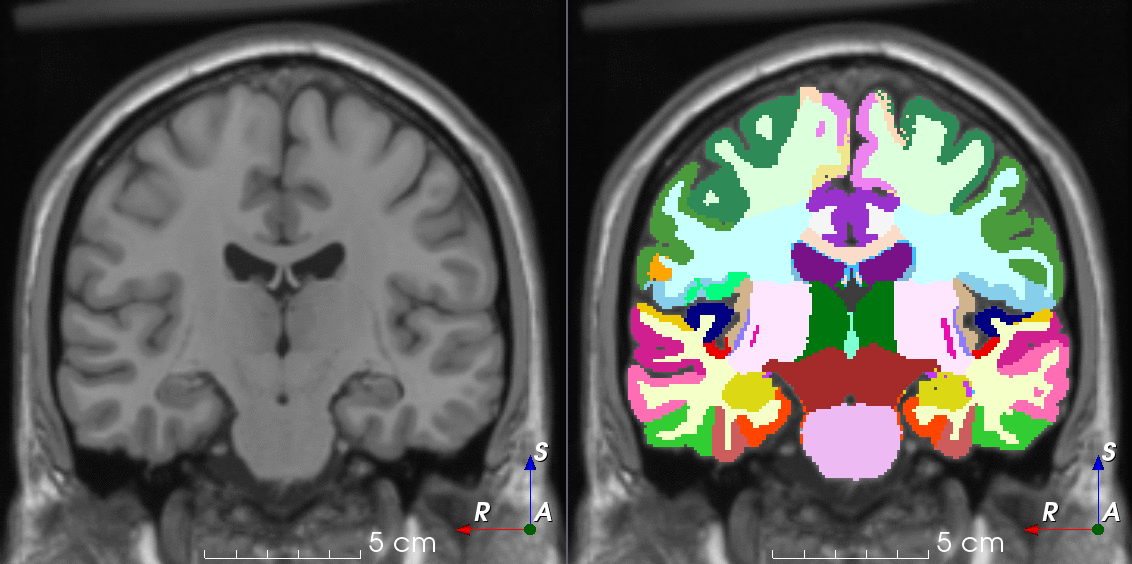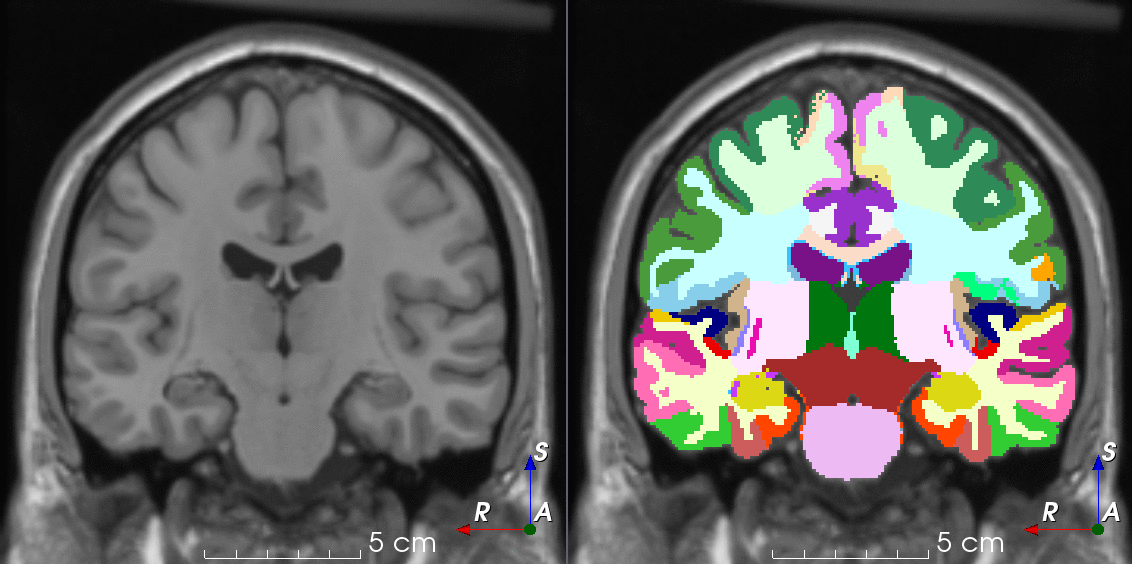
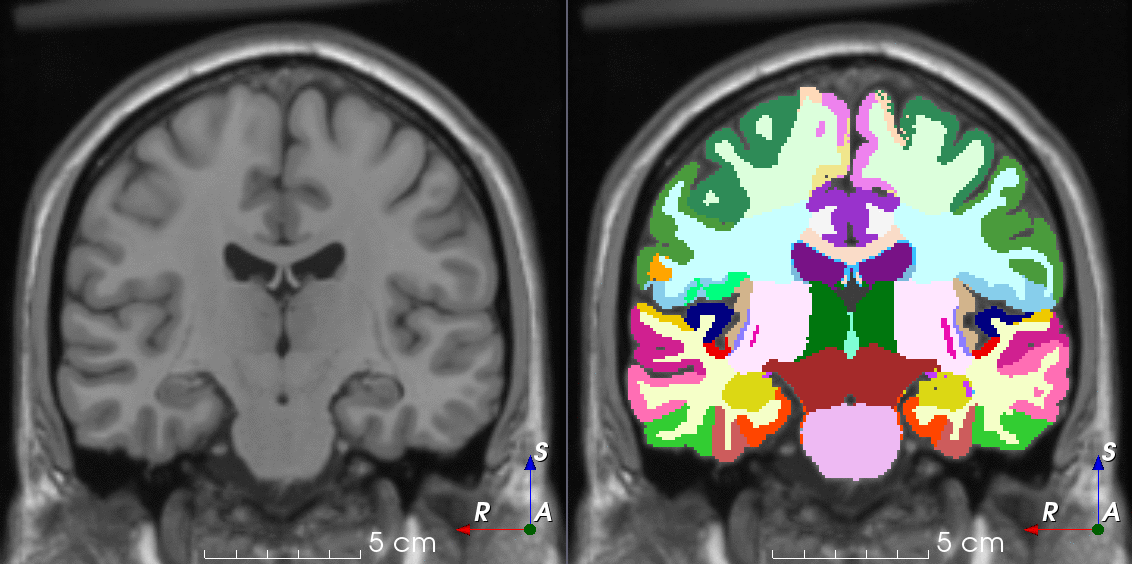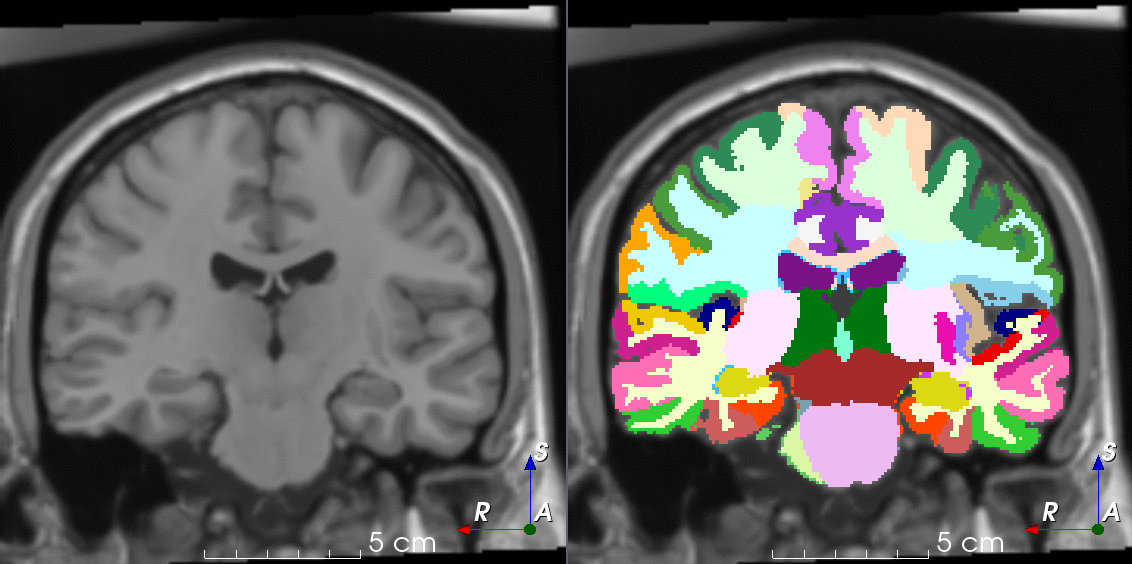
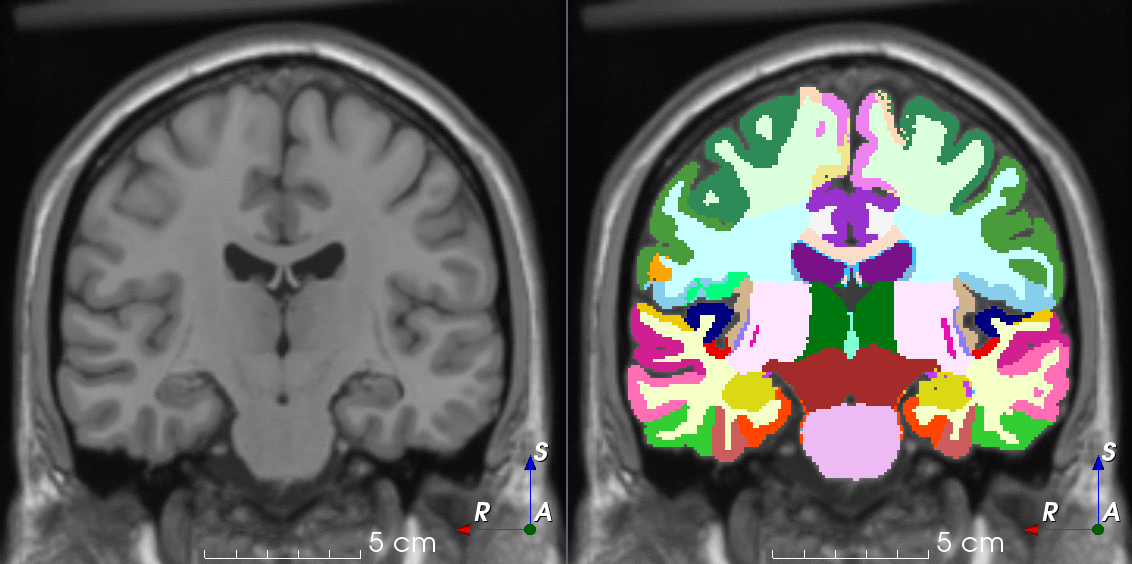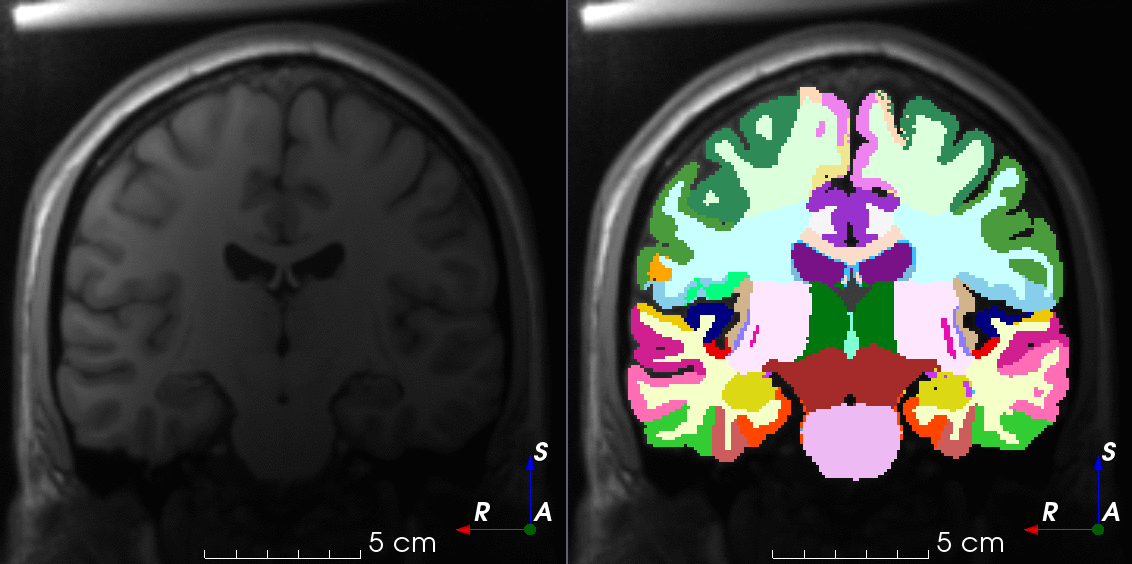
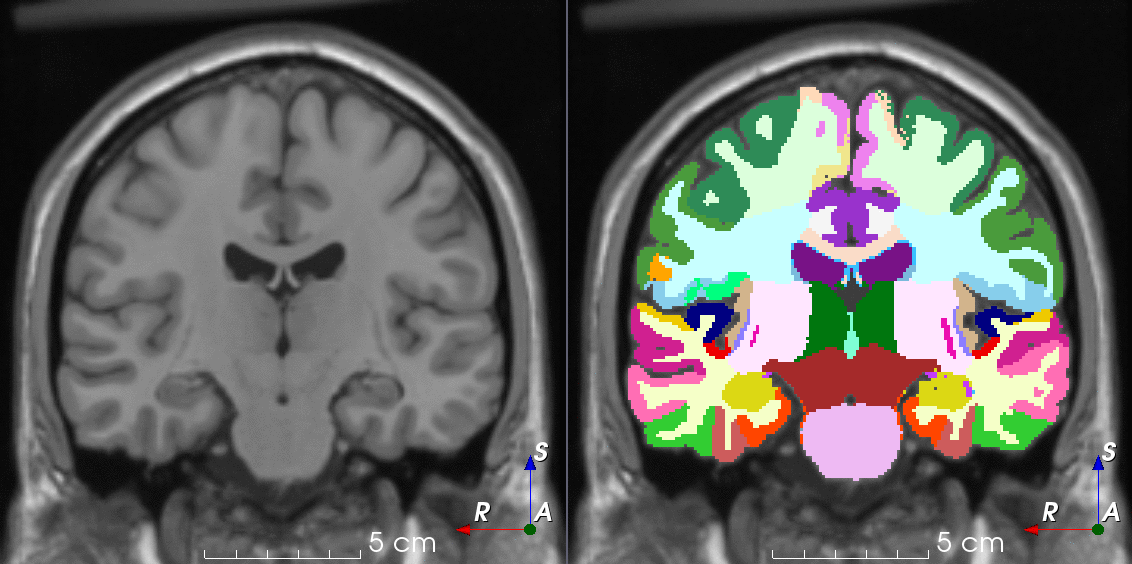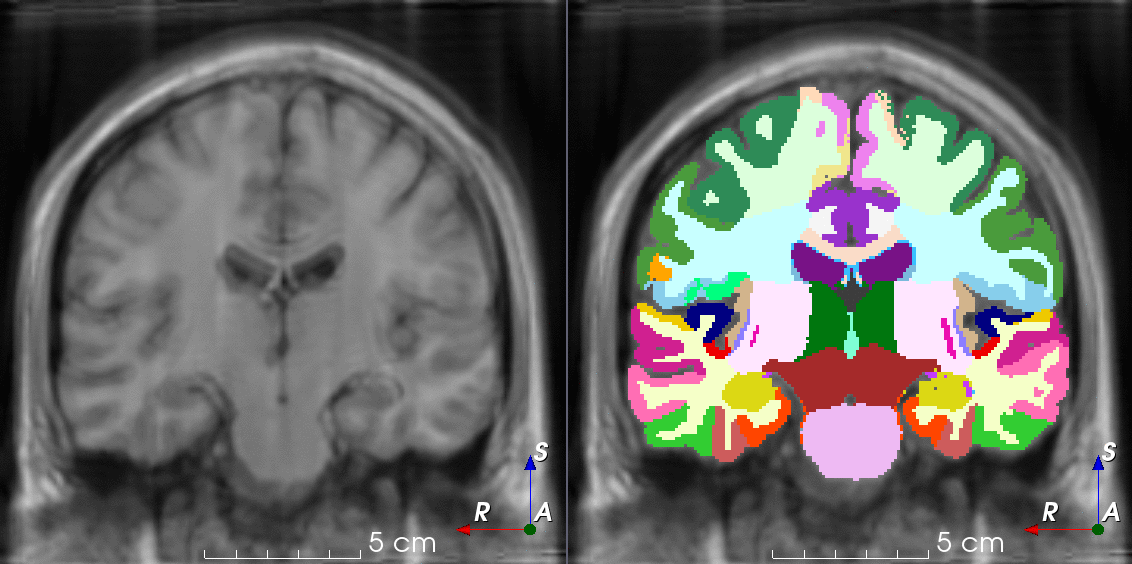
Tools like TorchIO are a symptom of the maturation of medical AI research using deep learning techniques.
Jack Clark, Policy Director at OpenAI (link).
| Package |



|
| CI |

|
| Code |


|
| Tutorials |
|
| Community |




|
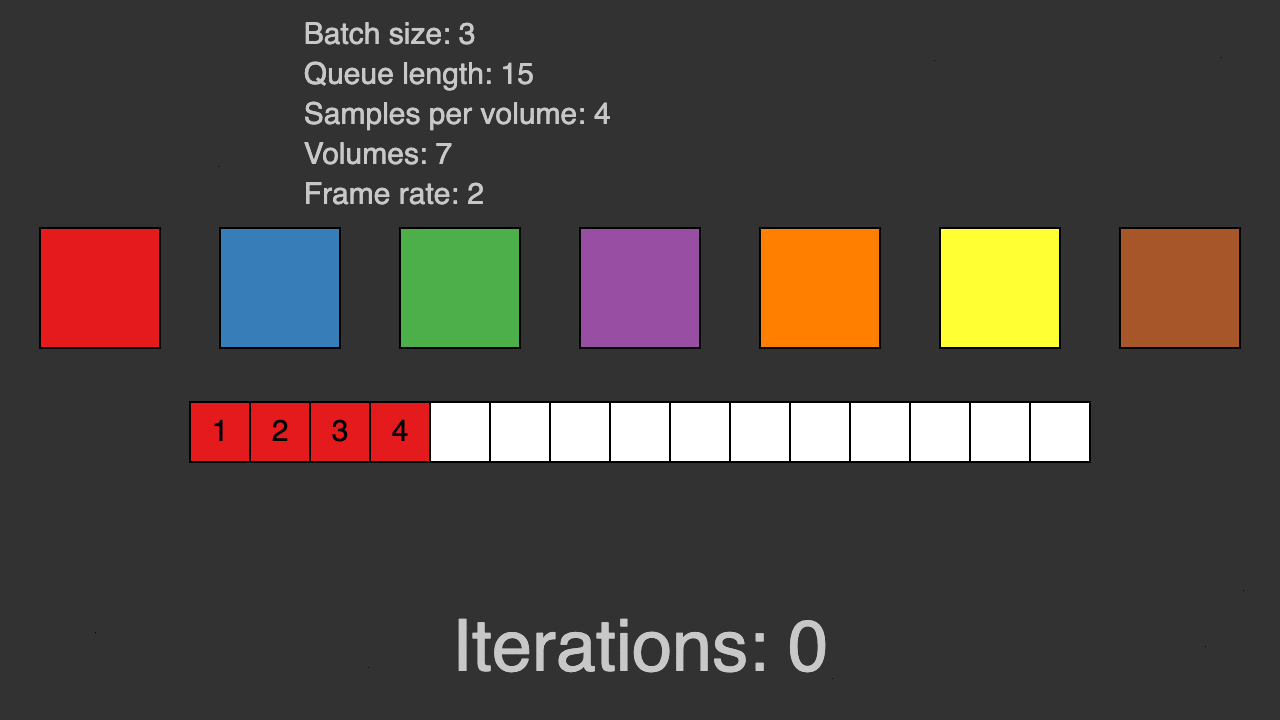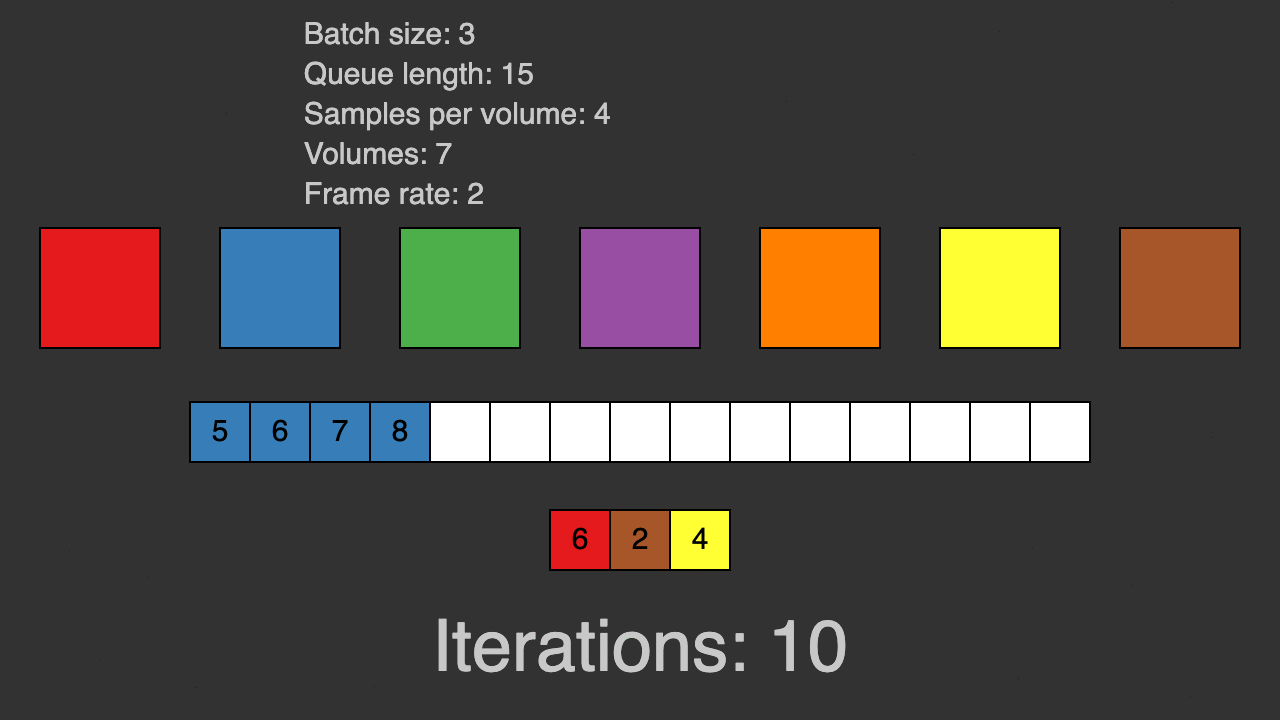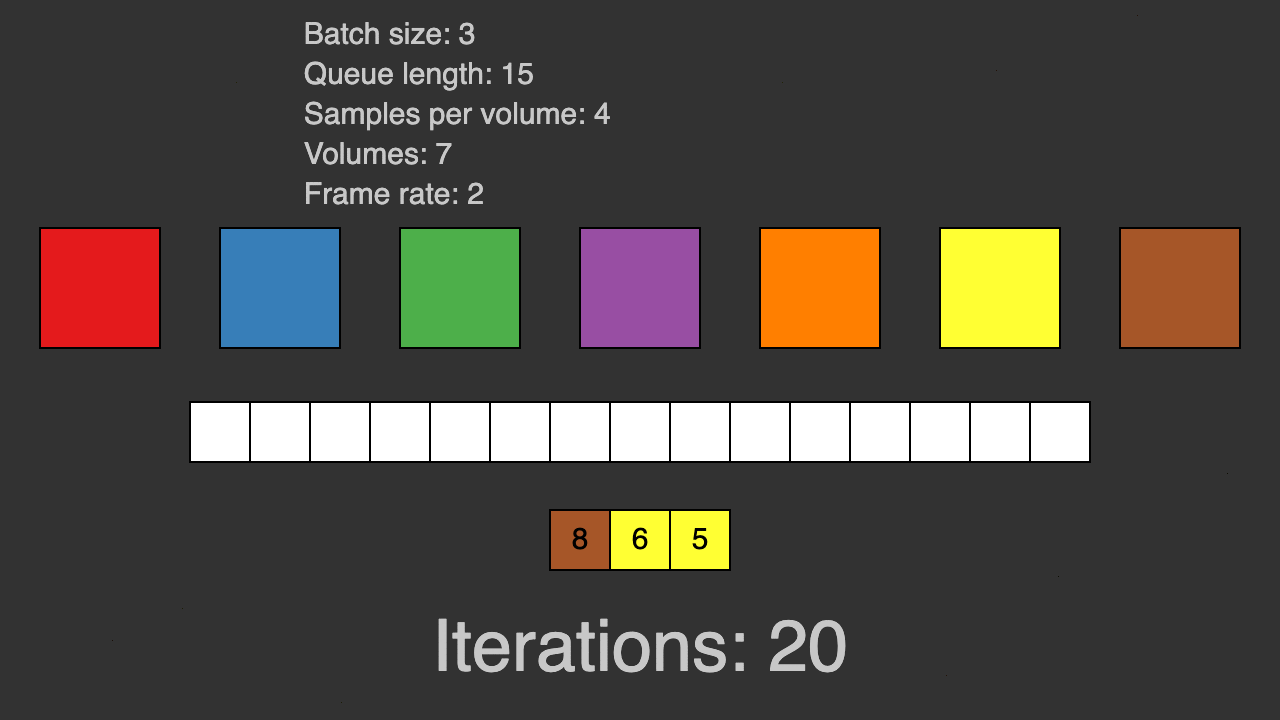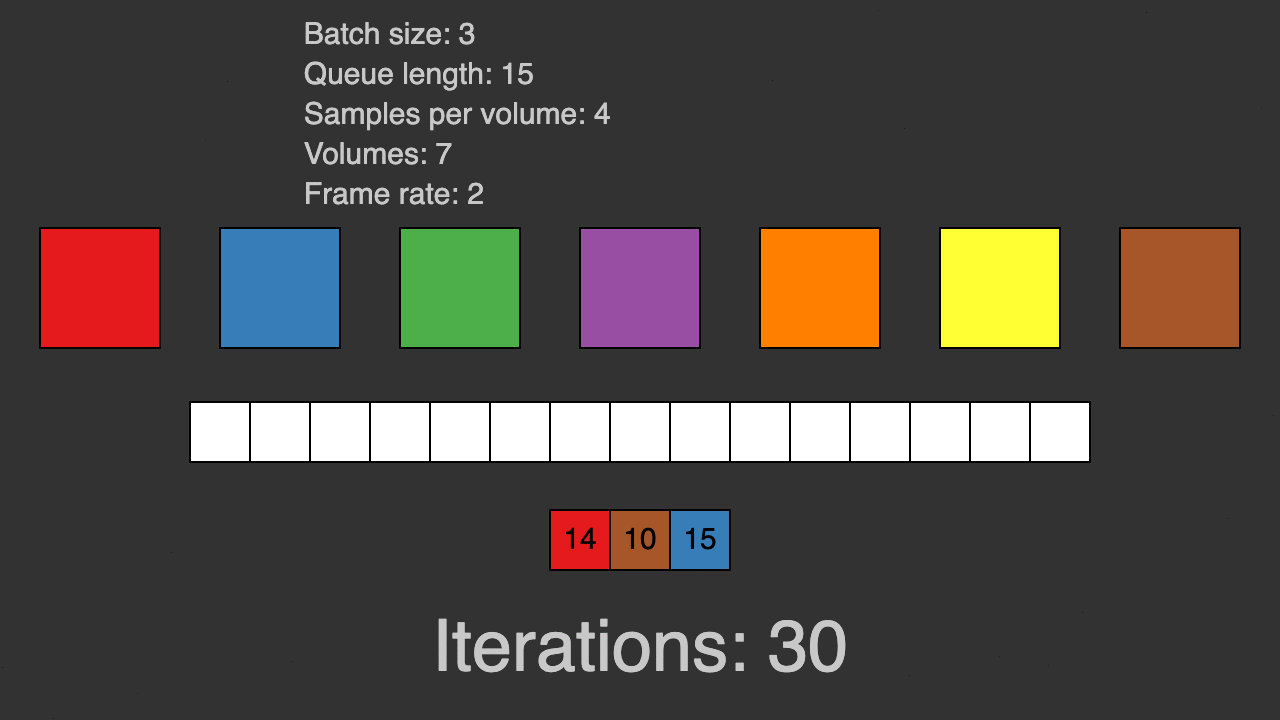
(Queue for patch-based training)
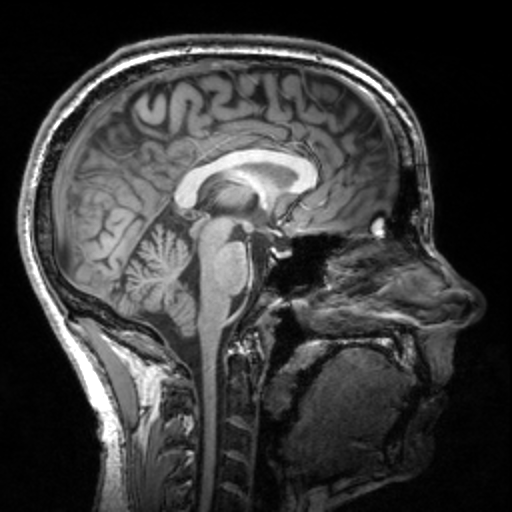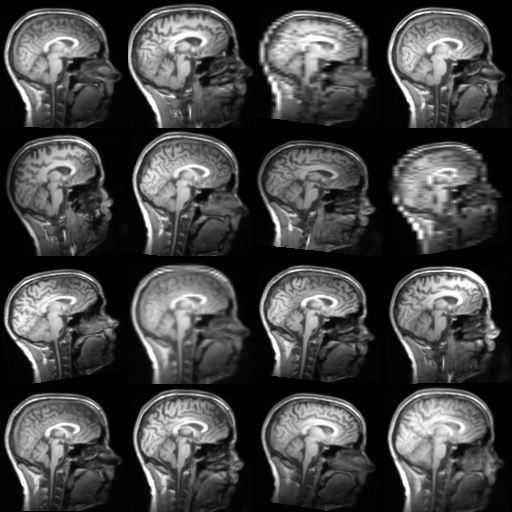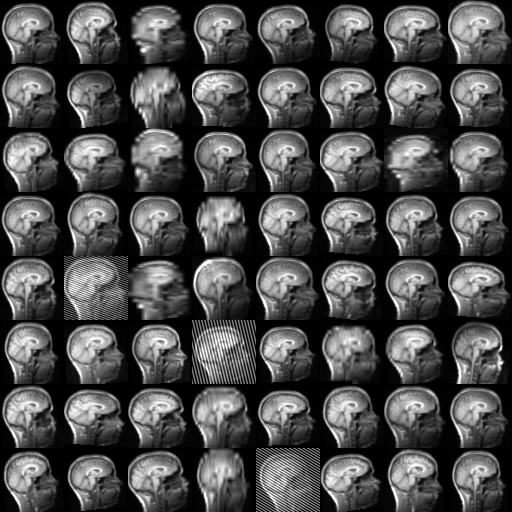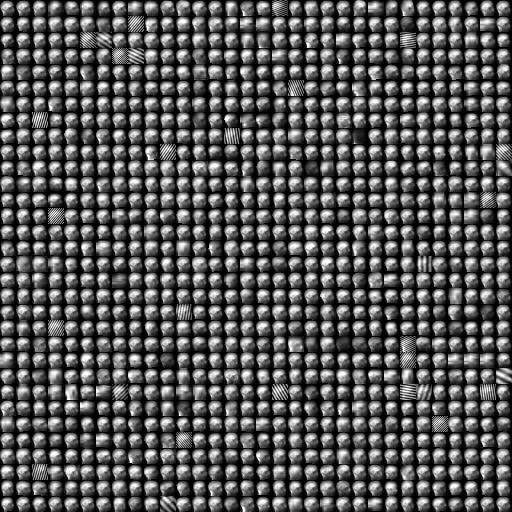
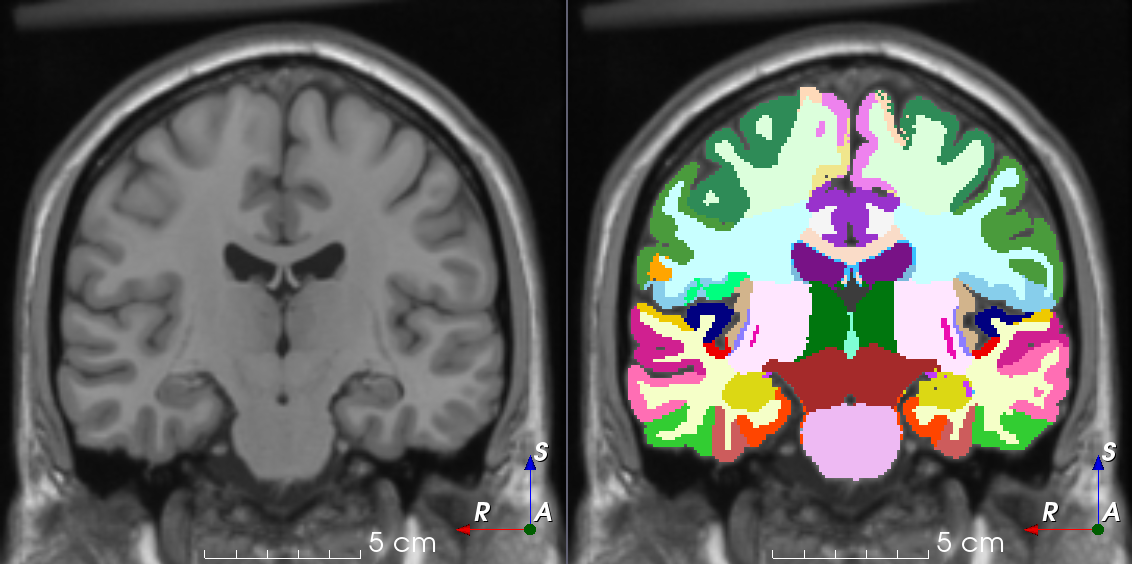
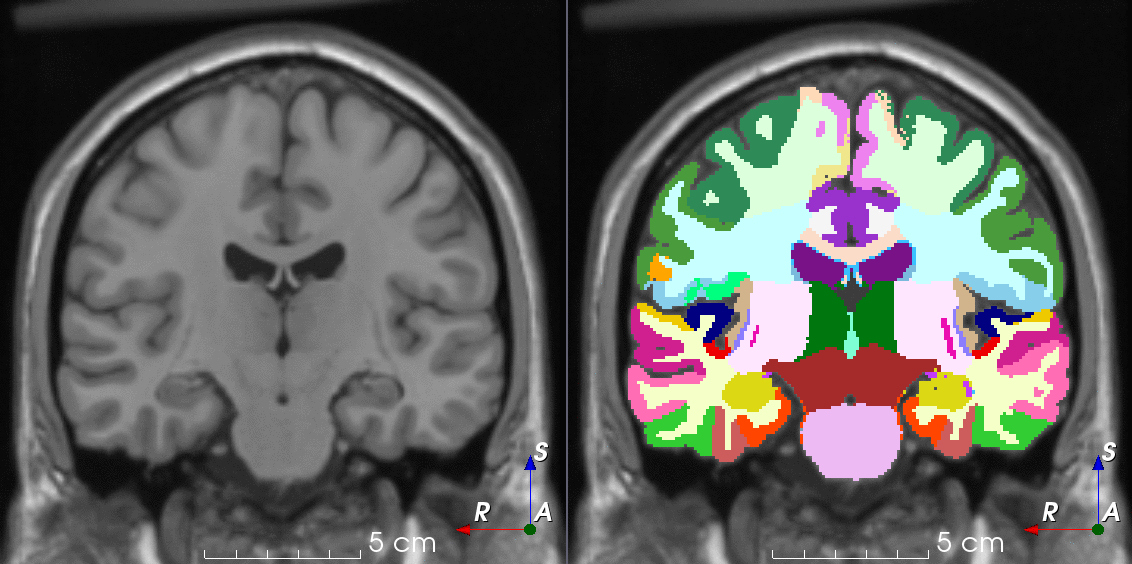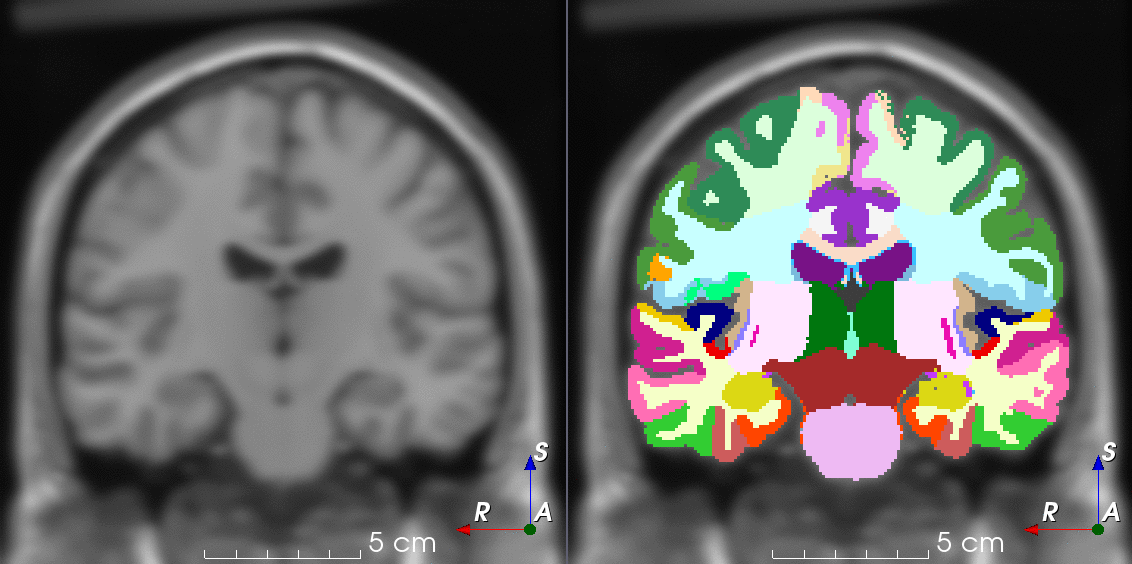
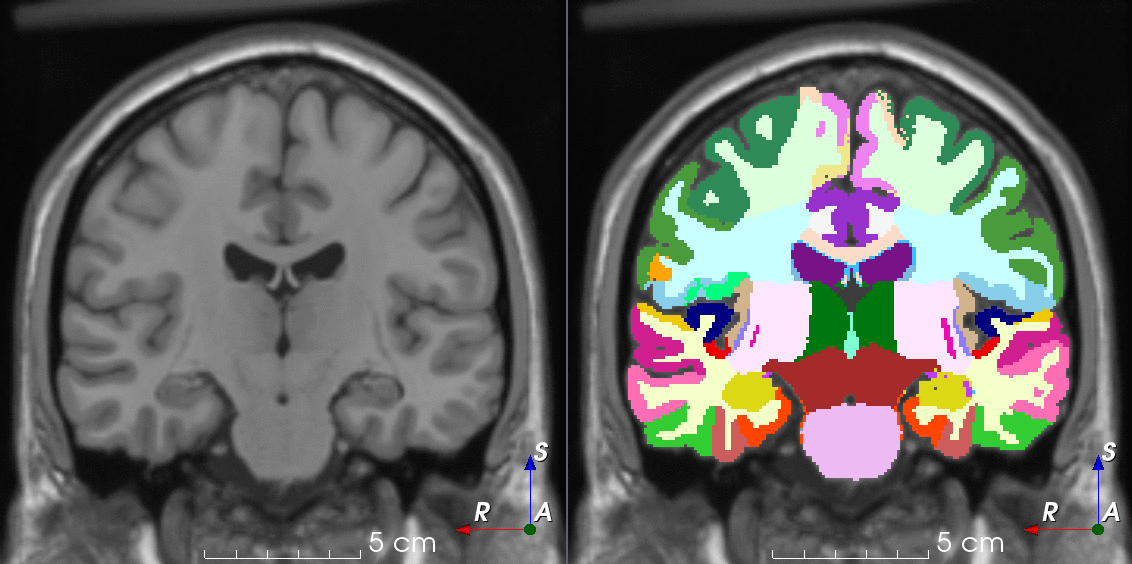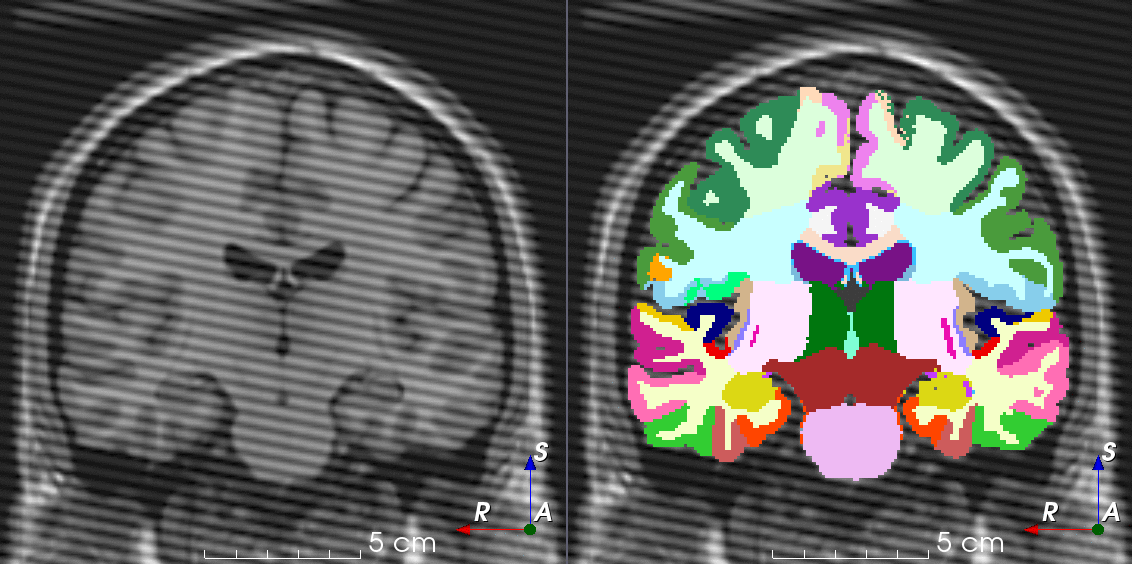
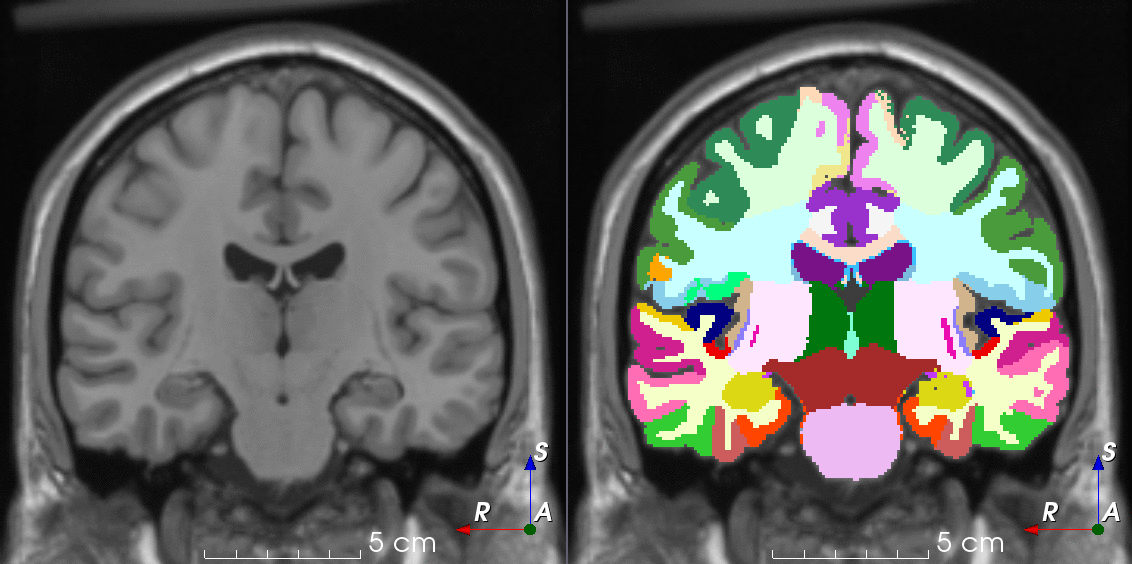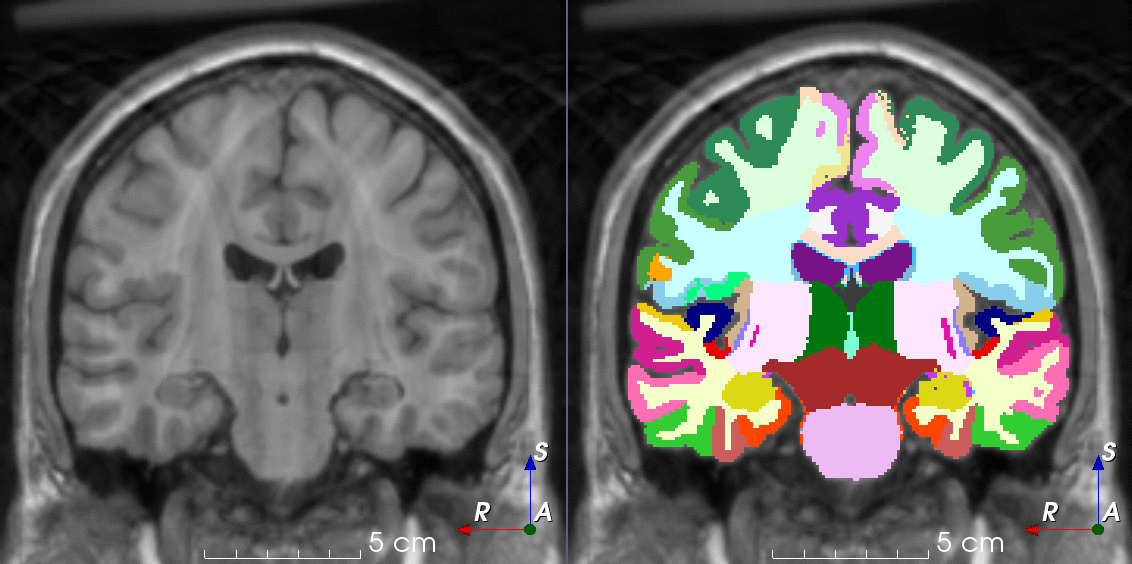
TorchIO is a Python package containing a set of tools to efficiently read, preprocess, sample, augment, and write 3D medical images in deep learning applications written in PyTorch, including intensity and spatial transforms for data augmentation and preprocessing. Transforms include typical computer vision operations such as random affine transformations and also domain-specific ones such as simulation of intensity artifacts due to MRI magnetic field inhomogeneity or k-space motion artifacts.
This package has been greatly inspired by NiftyNet, which is not actively maintained anymore.
If you like this repository, please click on Star!
If you use this package for your research, please cite our paper:
BibTeX entry:
@article{perez-garcia_torchio_2021,
title = {{TorchIO}: a {Python} library for efficient loading, preprocessing, augmentation and patch-based sampling of medical images in deep learning},
journal = {Computer Methods and Programs in Biomedicine},
pages = {106236},
year = {2021},
issn = {0169-2607},
doi = {https://doi.org/10.1016/j.cmpb.2021.106236},
url = {https://www.sciencedirect.com/science/article/pii/S0169260721003102},
author = {P{\'e}rez-Garc{\'i}a, Fernando and Sparks, Rachel and Ourselin, S{\'e}bastien},
}This project is supported by the following institutions:
- Engineering and Physical Sciences Research Council (EPSRC) & UK Research and Innovation (UKRI)
- EPSRC Centre for Doctoral Training in Intelligent, Integrated Imaging In Healthcare (i4health) (University College London)
- Wellcome / EPSRC Centre for Interventional and Surgical Sciences (WEISS) (University College London)
- School of Biomedical Engineering & Imaging Sciences (BMEIS) (King's College London)
See Getting started for installation instructions and a Hello, World! example.
Longer usage examples can be found in the tutorials.
All the documentation is hosted on Read the Docs.
Please open a new issue if you think something is missing.
Thanks goes to all these people (emoji key):
This project follows the all-contributors specification. Contributions of any kind welcome!