Overview | Features | Installation | Changelog | Documentation
OpenFL is a Python framework for Federated Learning. It enables organizations to train and validate machine learning models on sensitive data. It increases privacy by allowing collaborative model training or validation across local private datasets without ever sharing that data with a central server. OpenFL is hosted by The Linux Foundation.
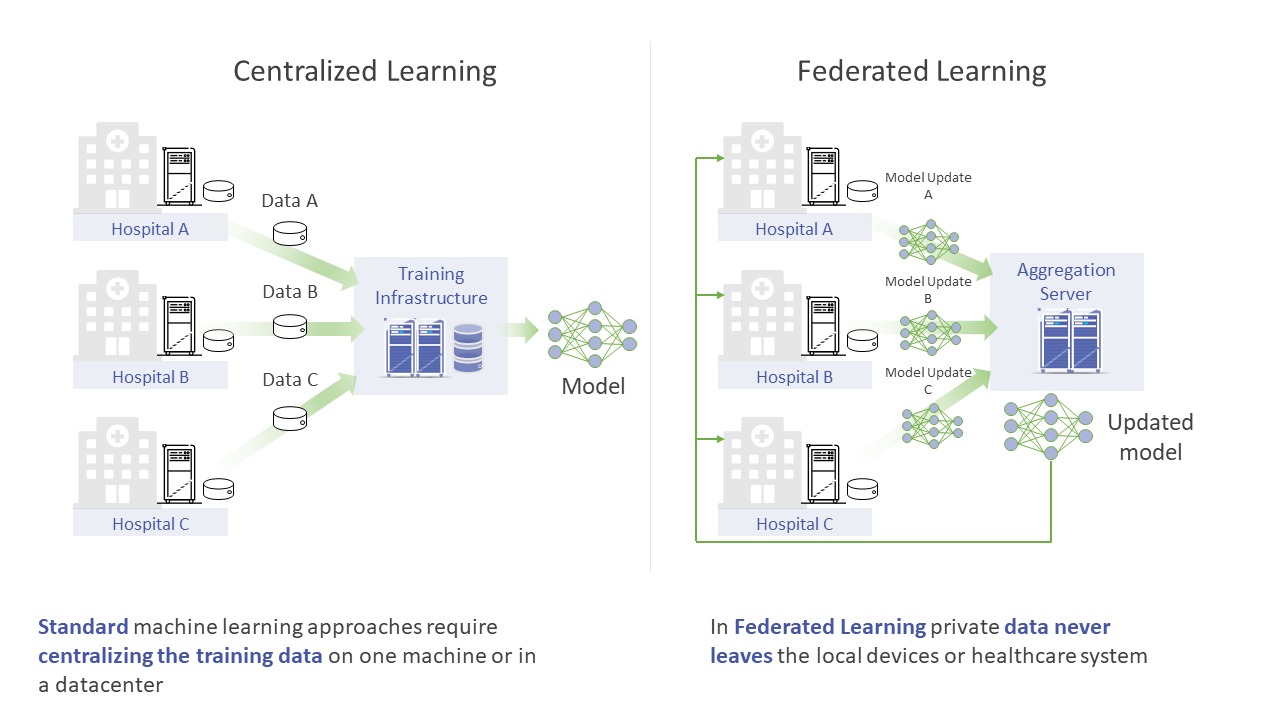
Federated Learning is a distributed machine learning approach that enables collaborative training and evaluation of models without sharing sensitive data such as, personal information, patient records, financial data, or classified information. The minimum data movement needed across a Federated Training experiment, is solely the model parameters and their updates. This is in contrast to a Centralized Learning regime, where all data needs to be moved to a central server or a datacenter for massively parallel training.
OpenFL builds on a collaboration between Intel and the Bakas lab at the University of Pennsylvania (UPenn) to develop the Federated Tumor Segmentation (FeTS) platform (grant award number: U01-CA242871).
The grant for FeTS was awarded from the Informatics Technology for Cancer Research (ITCR) program of the National Cancer Institute (NCI) of the National Institutes of Health (NIH), to Dr. Spyridon Bakas (Principal Investigator) when he was affiliated with the Center for Biomedical Image Computing and Analytics (CBICA) at UPenn and now heading the Division of Computational Pathology at Indiana University (IU).
FeTS is a real-world medical federated learning platform with international collaborators. The original OpenFederatedLearning project and OpenFL are designed to serve as the backend for the FeTS platform, and OpenFL developers and researchers continue to work very closely with IU on the FeTS project. An example is the FeTS-AI/Front-End, which integrates the group’s medical AI expertise with OpenFL framework to create a federated learning solution for medical imaging.
Although initially developed for use in medical imaging, OpenFL designed to be agnostic to the use-case, the industry, and the machine learning framework.
For more information, here is a list of relevant publications.
Install via PyPI (latest stable release):
pip install -U openfl
For more installation options, checkout the installation guide.
OpenFL supports two ways to set up a Federated Learning experiment:
-
TaskRunner API: This API uses short-lived components like the
AggregatorandCollaborator, which terminate at the end of an FL experiment. TaskRunner supports mTLS-based secure communication channels, and TEE-based confidential computing environments. -
Workflow API: This API allows for experiments beyond the traditional horizontal federated learning paradigm using a pythonic interface. It allows an experiment to be simulated locally, and then to be seamlessly scaled to a federated setting by switching from a local runtime to a distributed, federated runtime.
Note: This is experimental capability.
OpenFL is backend-agnostic. It comes with support for popular NumPy-based ML frameworks like TensorFlow, PyTorch and Jax which should be installed separately. Users may extend the list of supported backends if needed.
OpenFL supports popular aggregation algorithms out-of-the-box, with more algorithms coming soon.
| Reference | PyTorch backend | TensorFlow backend | NumPy backend | |
|---|---|---|---|---|
| FedAvg | McMahan et al., 2017 | yes | yes | yes |
| FedOpt | Reddi et al., 2020 | yes | yes | yes |
| FedProx | Li et al., 2020 | yes | yes | - |
| FedCurv | Shoham et al., 2019 | yes | - | - |
We welcome contributions! Please refer to the contributing guidelines.
The OpenFL community is expanding, and we encourage you to join us. Connect with other enthusiasts, share knowledge, and contribute to the advancement of federated learning by joining our Slack channel.
Stay updated by subscribing to the OpenFL mailing list: [email protected].
This project is licensed under Apache License Version 2.0. By contributing to the project, you agree to the license and copyright terms therein and release your contribution under these terms.
@article{openfl_citation,
author={Foley, Patrick and Sheller, Micah J and Edwards, Brandon and Pati, Sarthak and Riviera, Walter and Sharma, Mansi and Moorthy, Prakash Narayana and Wang, Shi-han and Martin, Jason and Mirhaji, Parsa and Shah, Prashant and Bakas, Spyridon},
title={OpenFL: the open federated learning library},
journal={Physics in Medicine \& Biology},
url={http://iopscience.iop.org/article/10.1088/1361-6560/ac97d9},
year={2022},
doi={10.1088/1361-6560/ac97d9},
publisher={IOP Publishing}
}